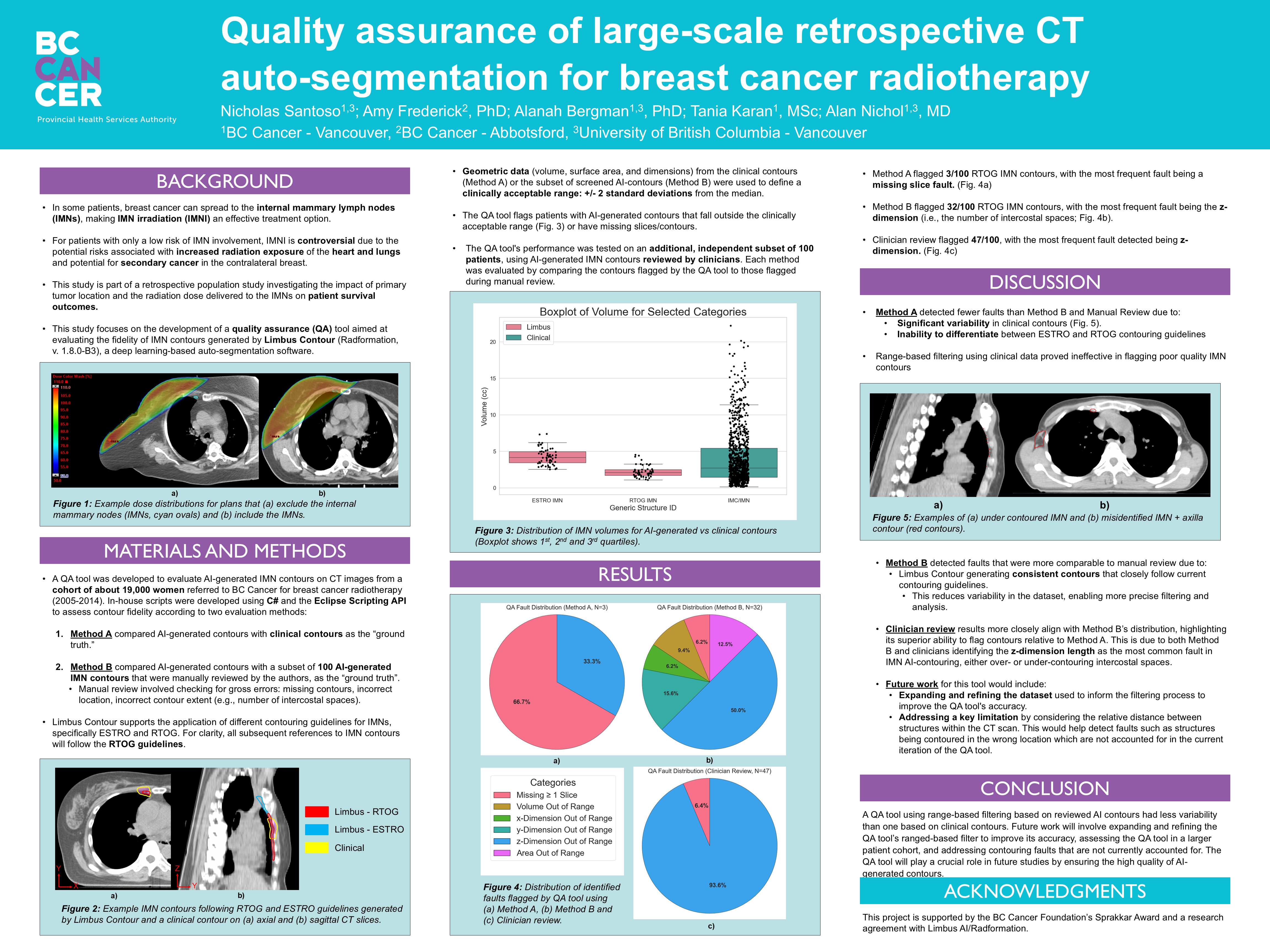
Background
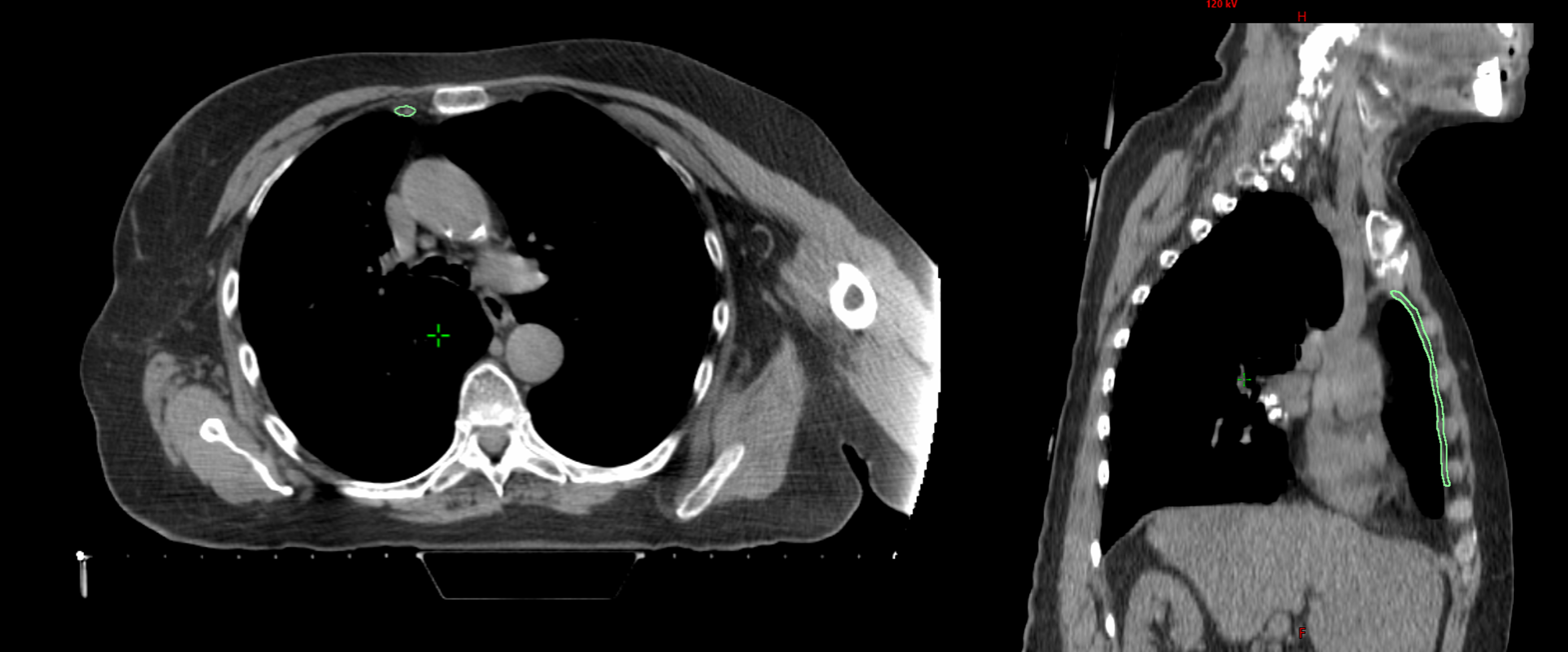
Breast cancer can spread to the internal mammary lymph nodes (IMNs), located near the center of the chest. When involvement is confirmed, IMN irradiation (IMNI) can improve outcomes; however, its use in low-risk patients remains controversial due to potential radiation exposure to the heart, lungs, and contralateral breast. This project contributes to a retrospective population study at BC Cancer investigating how primary tumor location and IMN radiation dose relate to survival outcomes. To support this research, a data pipeline is being developed to automate the import and segmentation of approximately 19,000 patient CT datasets (2005–2014) using Limbus Contour, a deep learning-based auto-segmentation software. My contributions include developing C# scripts for contour transfer and a QA system to evaluate the accuracy of AI-generated contours for future pipeline integration.
1.0 Project Objectives
- Develop a QA pipeline to assess quality of AI-generated IMN contours.
- Establish a reliable ground-truth set for evaluation.
- Support retrospective research into the effects of IMN irradiation on survival outcomes.
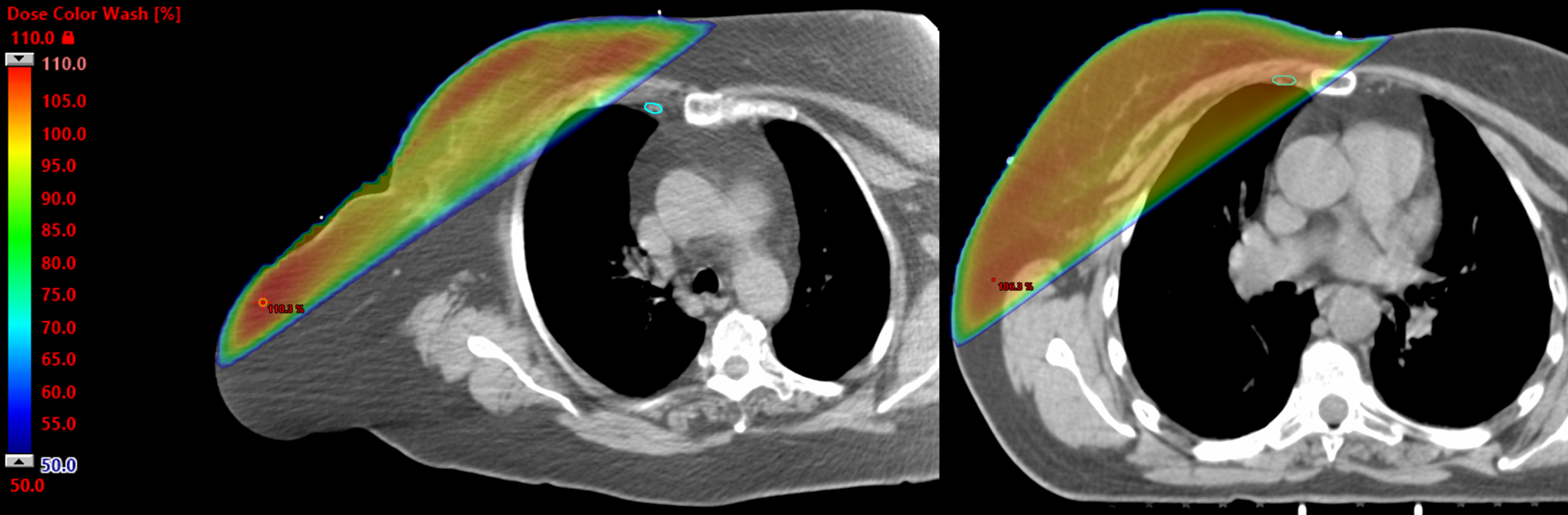
- Clinical IMN contours were inconsistent due to varying guidelines (RTOG vs ESTRO) and incomplete slices.
- AI-generated contours required systematic QA to identify errors before use in dosimetric studies.
2.0 My Contributions
- Developed contour transfer and QA scripts in C# using the Eclipse Scripting API.
- Designed statistical filtering logic (Median ± 2SD, IQR, Min/Max) for quality flagging.
- Compared performance of different thresholds using a reviewed subset of 100 patient cases.
- Refined the algorithm to balance false positive and false negative rates.
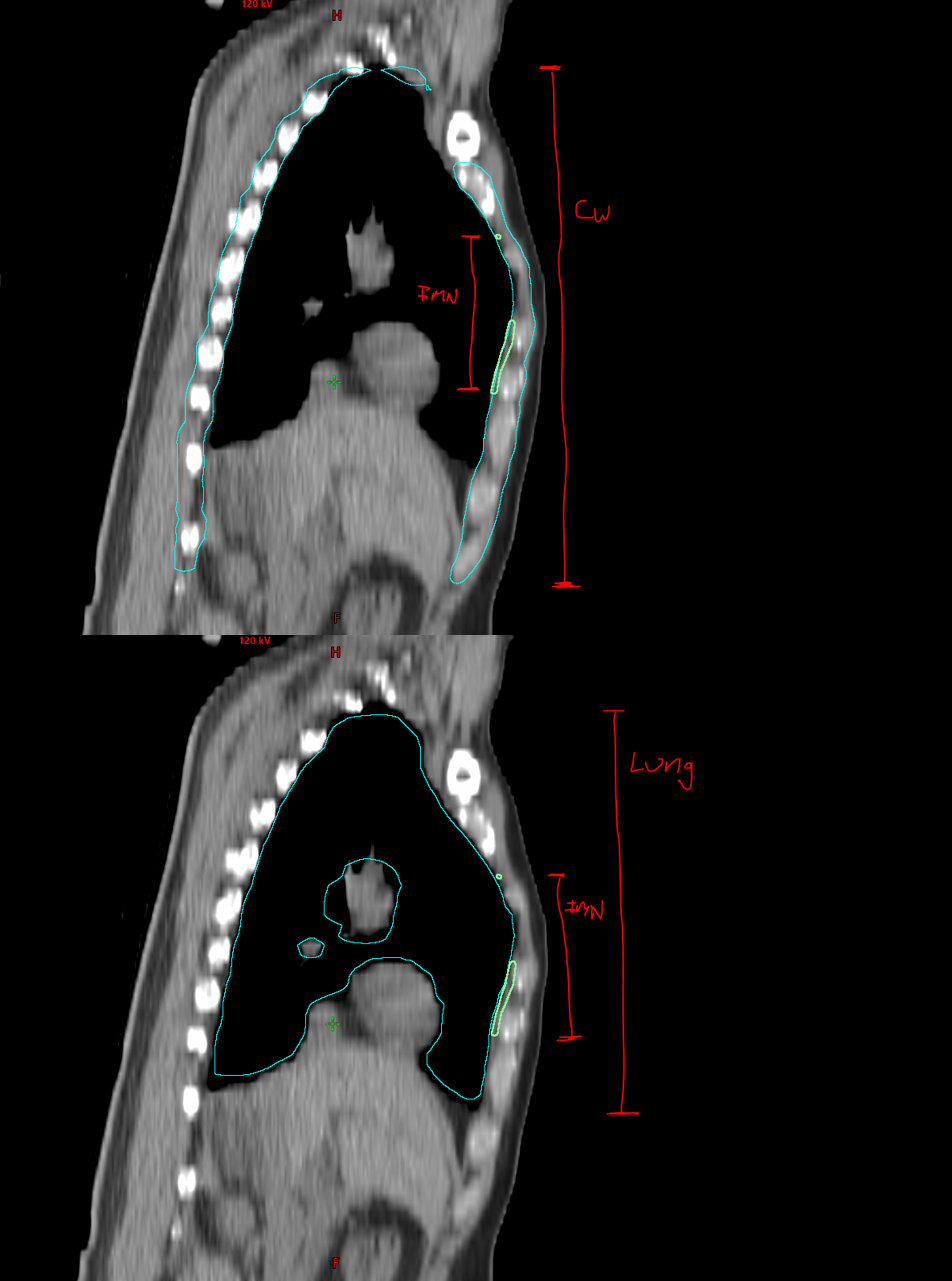
- Introduced normalized ratio metrics (IMN:Lung, IMN:Chest Wall) to reduce anatomical variability bias.
- Implemented relative ratio metrics between the IMN and reference organs to enhance filter reliability and account for anatomical variability across patients.
- Contributed to the research poster and presentation at the BC Cancer Summit 2024.
3.0 Methodology
3.1 Implementation
In-house scripts were written in C# using the Eclipse Scripting API (ESAPI) to analyze contour quality. The QA tool flagged patients with contours outside of statistically defined ranges (median ± 2SD, Min/Max, IQR) or with structural faults such as missing slices.
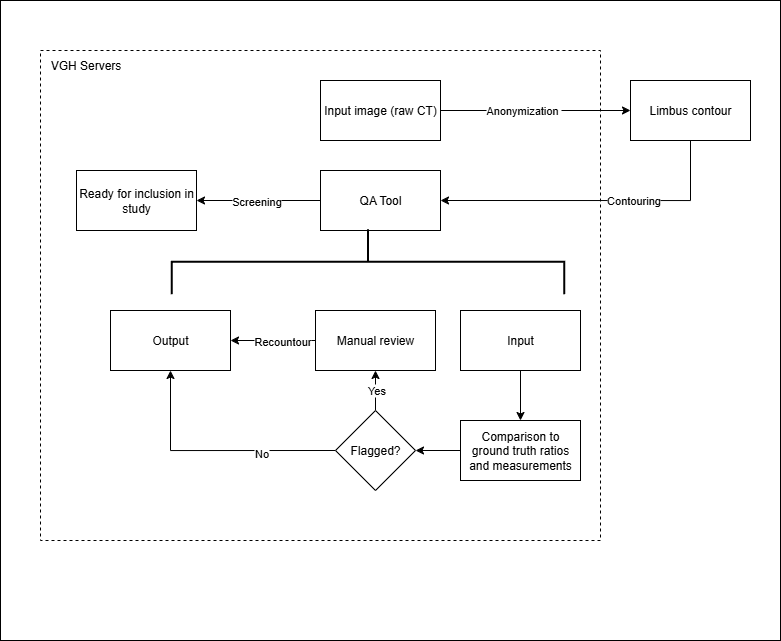
3.2 Evaluation Approaches
- Method A: Compared AI contours against clinical contours. This was unreliable due to inconsistent guidelines (RTOG vs ESTRO) and incomplete slices.
- Method B: Compared AI contours to a reviewed subset of 100 Limbus-generated contours (RTOG guideline), providing a consistent baseline.
3.3 Technical Challenges
One of the main challenges was that filtering contours based purely on their absolute length along the z-axis could inadvertently exclude patients with atypical anatomy, such as very tall or very short individuals. To address this, we instead used ratios (such as IMN:Lung and IMN:Chest Wall) to normalize contour dimensions relative to each patient’s anatomy.
4.0 Key Results
| Method | Contours Flagged (n=100) | Most Common Error |
|---|---|---|
| Method A (Clinical) | 3 | Missing slices |
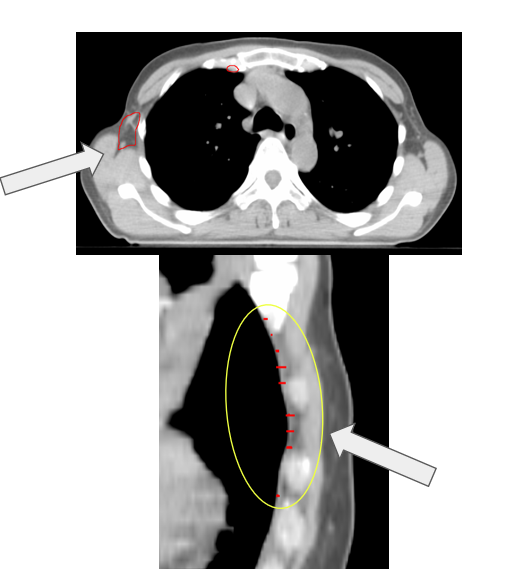
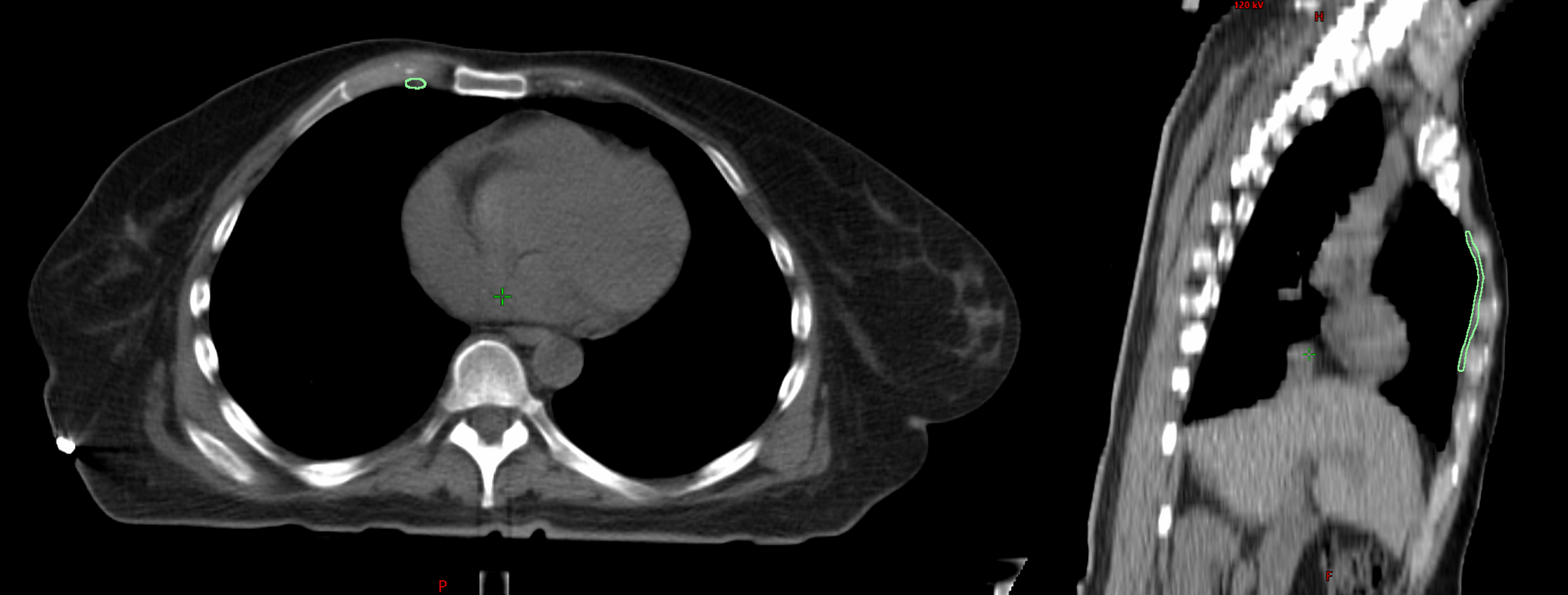
| Method B (AI-reviewed subset) | 32 | Z-length errors |
| Clinician Review | 47 | Z-length errors |
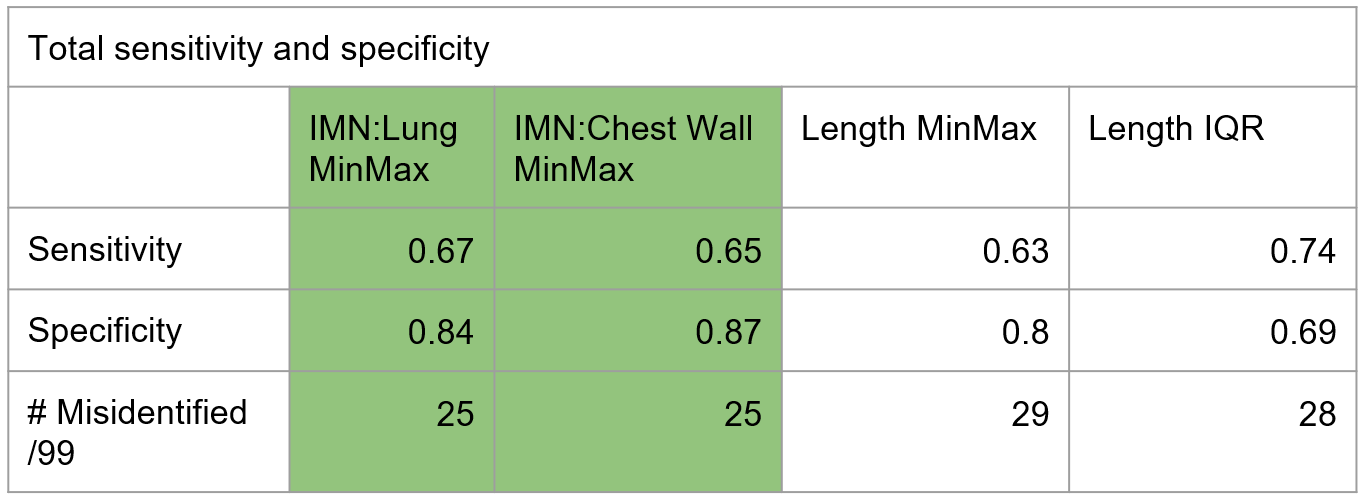
5.0 Findings & Discussion
Clinical contours were too inconsistent to serve as a reliable ground-truth. The reviewed subset of Limbus contours was more effective for QA. The most common failure mode was incorrect Z-dimension contouring. Absolute length metrics introduced patient-size bias, making normalized ratios a better approach.
6.0 Tools & Technologies
C# Eclipse Scripting API (ESAPI) Medical Imaging Deep Learning QA Data Normalization
7.0 Next Steps
- Expand the dataset used for defining clinically acceptable ranges.
- Incorporate anatomical context (e.g., relative placement to chest wall or lungs).
- Integrate QA directly into the pipeline for real-time flagging of errors.
- Validate the tool across larger and independent patient cohorts.
8.0 Acknowledgments
This project was supported by the BC Cancer Foundation’s Sprakkar Award and a research agreement with Limbus AI/Radformation. Collaborators include Amy Frederick (PhD), Alanah Bergman (PhD), Tania Karan (MSc), and Alan Nichol (MD).
9.0 Reflection
This project taught me how to bridge machine learning with clinical workflows, emphasizing the importance of quality assurance in large-scale AI studies. It also deepened my understanding of radiotherapy planning and the practical challenges of integrating AI into healthcare.